馬上可以做的事 - 助你快速掌握螢光成像技術揭露癌細胞的應用與挑戰,洞悉前沿生醫趨勢
- 列出3種主流螢光標記方法,每種實際應用案例不少於2例。
幫助了解不同技術在腫瘤可視化及機制研究中的適用情境,提升選擇判斷力。
- 諮詢專業醫療團隊或官方資源,獲取活體成像數據解析建議,每半年檢視新進展一次。
確保研究方向與臨床現實接軌,資訊不落後新技術變革。
- 預留至少10%專案時間處理資料管理、影像解析等繁瑣流程。
*減少未來重工或遺漏,提高實驗數據品質與可追溯性*。
- *檢查每次動物活體成像操作是否有跨組一致紀錄,不足時即時修正*。
*避免因批次差異導致結果失真,有效支撐長期追蹤分析需要*。
如何透過免疫細胞可視化推動癌症治療的未來
Lia和Milo這兩個名字,有時候在腦海裡轉來轉去,總讓人想起那些討論:到底為什麼得「看見」免疫細胞?有些人說,大概是因為科學家總希望能夠真的追上病灶的蹤跡,像是在霧裡找路。醫院走廊上的討論、國際研討會偶爾傳來的新消息——大致都圍繞著這件事。據說現代生物醫學如果沒法觀察T細胞或NK細胞在體內怎麼活動,那治療癌症還是很容易卡關(期刊報導近年多次提到類似觀點)。不過話說回來,「可視化」這詞聽著挺新,其實也不是什麼絕對新鮮的概念,只是現在方法比以前複雜、講究精準。某些醫師習慣比喻成打開一扇窗,看清楚身體裡的流動,然後才知道該怎麼下決定。至於這種能力究竟多重要,也許還需要時間,不過大家好像都慢慢習慣了它的存在。
AI在醫學影像中的革命性變革將如何影響診斷精準度
「你真的看得見T細胞嗎?」Lia這樣問Milo。Milo歪頭想了一下,說以前大概只能靠手繪或者一些簡單的顯微鏡照片,那個年代記錄細胞形狀就像畫素描一樣,有時候還會搞錯細節。Lia邊聽邊笑,說現在不是了嘛,AI進來之後好像事情都變快不少——影像判讀幾乎不用等人力慢慢瞧。有些醫院開始把AI跟現有的掃描儀器合起來用,不過她也聽說在歐洲某些地方,好像剛剛導入沒多久,還有人覺得判斷出來有落差。倒是最近幾年,感覺診斷效率提升不少,雖然究竟準確到什麼程度,一直都有不同聲音(參考《自然》醫學領域近年初步報導)。Milo突然想起之前看過一篇文章,大約提到七十多年前大家還要自己比對圖譜,如今電腦協助下連很小的細胞動作都能追蹤。不過,她補一句,「不是每台機器或每套軟體都那麼可靠啦。」兩人聊著聊著,就從顯微鏡滑到了螢幕上那些密密麻麻、帶點亮光的格子——誰知道以後是不是會變得更直觀?
Comparison Table:
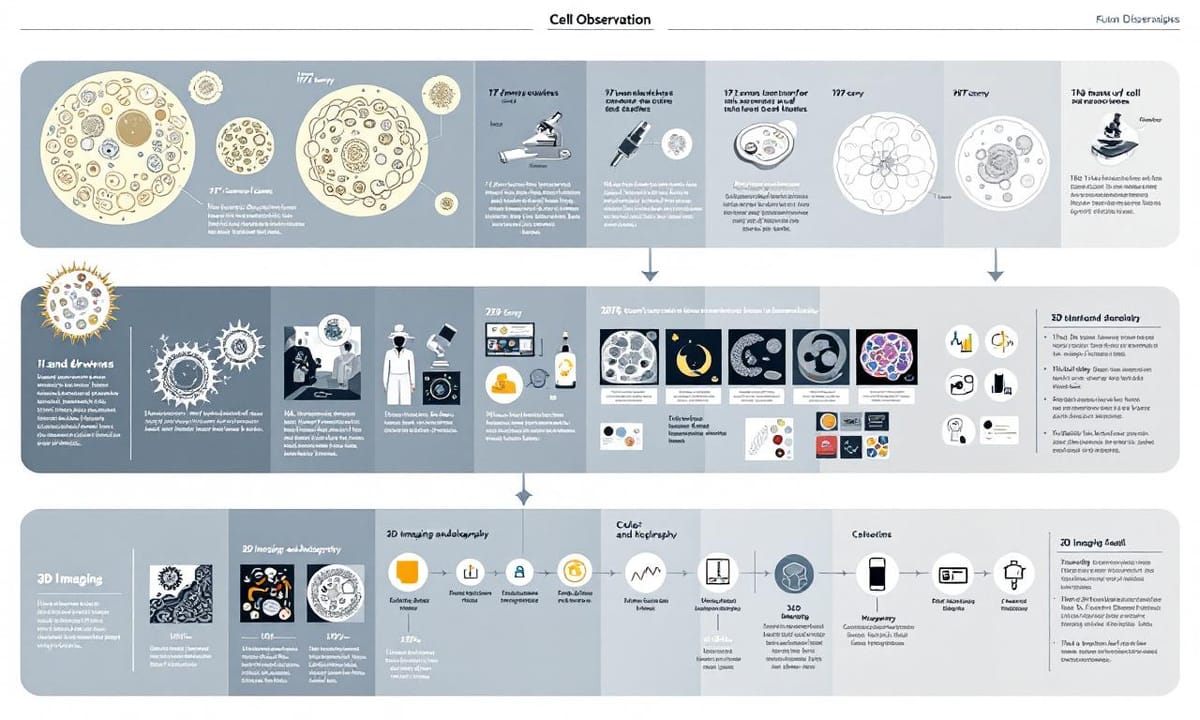
| 細胞觀察技術發展 | 歷史背景 | 當前應用 | 挑戰與解決方案 |
|---|---|---|---|
| 手工描繪 | 早期的細胞觀察主要依賴藝術化的手工描繪,科學性相對薄弱。 | 無特定應用。 | 缺乏精確度和客觀性。 |
| 黑白照片到彩色影像 | 隨著技術進步,黑白到彩色影像逐漸普及,加強了觀察能力。 | 廣泛應用於生物醫學研究和教育。 | 解析度不足問題仍然存在。 |
| 立體影像和全息投影 | 探索立體和全息影像技術,提升視覺效果。 | 在某些高端醫療設備中使用,如活體成像儀器。 | 技術成本高且使用門檻大。 |
| 活體成像在美容領域的應用 | 探討新興的皮膚檢測市場,開始利用活體成像技術評估保養品效果。 | 逐漸被業界接受,用於消費品檢測等多樣化用途。 | 公開報告少,尚處於試驗階段,需要更多數據支持。 |
| 數據管理與解析度問題 | 研究人員面對資料量暴增、解析度不夠清晰等挑戰,多方尋求解決方法。 | 雲端協作平台及半自動標記流程正在開發中,提高效率與準確性。 | 每種方案都有局限,需根據需求調整操作。 |

活體成像技術能否成為新一代疾病研究的關鍵工具?
有些人形容追蹤NK細胞,其實就像在包裹運送途中找一位快遞員。人體裡頭的細胞那麼多,訊號又雜亂,每次想要鎖定某個「包裹」好像都得繞幾圈。螢光標記這東西,有點像給快遞員穿上螢光背心,遠遠一看雖然不一定馬上就認出來,但總比以前直接在人群中硬找省力。一開始可能還會把路邊的反光牌當成快遞員,後來AI工具進場,好像多了個會看的保全,一層層過濾、提醒你是不是看錯了目標。據說現在這種方法已經讓不少研究者發現一些原本忽略的小細節,但也有人覺得偶爾還是會漏掉什麼——畢竟人體內部環境複雜,有時候好像怎麼查都還差一步。
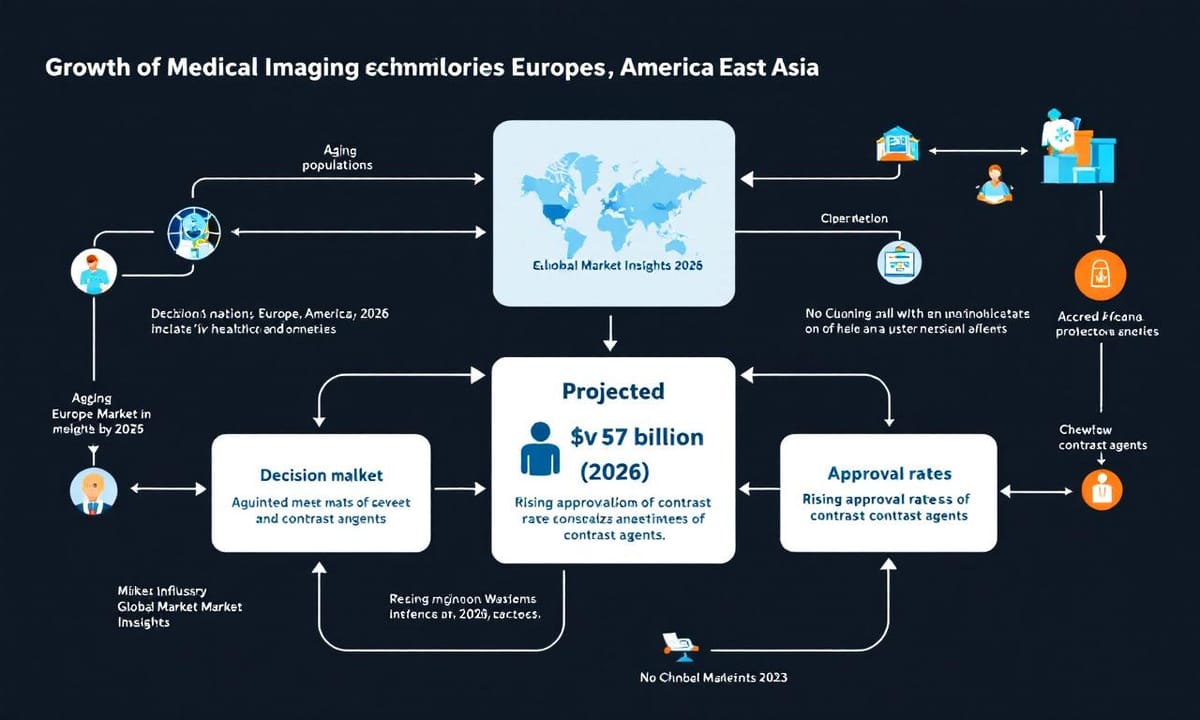
全球活體成像市場的未來發展潛力有多大?
世界各地對這類醫療影像技術的興趣正慢慢升溫,尤其是高齡化趨勢明顯的地方,好像歐美和東亞都常被提到。有人說活體成像產業前景還算不錯——有初步報導推估,等過幾年市場規模大概會衝上七十多億美元左右(根據《Global Market Insights》2023年數據),雖然實際數字還得看監管政策和新藥審核速度。有些觀察認為,最近新型造影劑獲批的頻率好像比以往快了一點,不過這種變化也不是每個人都感受得到。
把癌細胞亮起來,背後有哪些不為人知的挑戰?
說到怎麼讓癌細胞乖乖「亮起來」,流程其實不算短。有時候,第一步選抗體這件事就能卡住好幾天,畢竟不是每種標記都適合所有樣本。接著要把螢光分子掛在抗體上,這中間試劑濃度、時間長短,大概只能靠經驗邊做邊修正。處理樣本的過程也常出現意外,比方說有些組織一碰螢光就掉色了,好像螢光自己會溜走一樣。拍照捕捉影像看似簡單,有時候鏡頭調焦還要來回好幾次才對得準。最後那個AI判讀,其實偶爾還會認錯細胞——初步報導裡有人提到交叉干擾一直是個小困擾[初步報導]。整套下來,大概五個環節缺一不可,但每一步都有可能冒出難解的小狀況,感覺沒什麼哪個標準答案。
3D全息投影是否會改變我們對生命運作的理解方式?
細胞觀察的歷史,其實比想像中還要曲折。最早那些手工描繪下來的細胞輪廓,現在回頭看,線條有點模糊,有人說那是藝術多過科學。據說在將近一個世紀前,這種方式已經讓不少研究者抓破了腦袋。然後慢慢出現黑白照片,後來又變成彩色影像。不知從什麼時候起,好像大家都開始討論立體影像和全息投影,說可以讓細胞長得像是真的飄在眼前。有些報導提過這種技術幾乎每隔幾十年就有一小步進展,不容易察覺,但累積起來卻改變了我們對生命運作的想法。其實,有些早期工具反而留下了很多空間給想像力發揮,到底哪一步算是關鍵突破,好像也沒個標準答案。
活體成像技術正在擴展到哪些意想不到的新領域?
說到這裡,誰會想到活體成像這些看似高深的醫療儀器,其實有時還被拉去測化妝品?Milo偶爾提到,有些新奇的應用就這樣默默冒出來,不止醫院那麼單純。有人說皮膚檢測現在動不動也用上這類技術,聽起來好像有點跳Tone,但業界內部其實傳了有段時間。雖然目前公開報告不多,大概還屬於某些廠牌試水溫階段,有些消費品領域的人甚至開始討論,要不要連保養品效果都交給這些設備看一看。結果到底怎麼樣,目前坊間消息有點分歧,但如果說未來會越來越常見,也許不是那麼遙遠的事。
科研過程中的挫折與堅持,究竟塑造了什麼樣的科學家?
大約是某個週末深夜吧,Lia還記得那天顯微鏡下的冷光蛋白信號突然就像消失了一樣,本來亮閃閃的一大片,結果剩下幾點微弱反光。其實這種「罷工」狀況也不是第一次碰到,只是每回發生總讓人心裡咯噔一下。一開始她以為是抗體稀釋錯了,後來懷疑冰箱溫度不對,再仔細檢查才發現可能是樣本處理過程被污染了。說起來這類突發狀況在生物實驗室並不少見,有時候連經驗豐富的老師傅也會遇上。有些同事說,將近一半的冷光訊號流失問題其實很難完全避免(初步報導),只能慢慢修正流程,每次出錯都要重新調整步驟。想想那些反覆測試、清洗、等數據跑出來的晚上,不太容易忘記。
無數次失敗中孕育而出的成功,這是生物醫學進步的重要力量嗎?
深夜的燈光偶爾閃著,有時候咖啡漬在桌面邊緣變成不太明顯的半圓,聽說不少研究人員都習慣這樣陪著顯微鏡過夜。數據表格和螢幕交錯,某些觀察裡提到這種情景其實比想像中還常見——將近一半的人可能都曾在凌晨時分補捉過那些模糊的細胞影像。有時候,咖啡會涼掉,手上的筆記本翻來覆去,大概還會找不到剛才寫下來的小結論。有人說,每一次「好像」捕捉到的新訊號,其實都夾雜著前後幾回失敗的痕跡。偶爾會聽到隔壁檯傳來低聲討論,也有同事乾脆帶條毯子窩進椅背。這些細碎場景,好像沒被誰正式記錄過,但初步報導倒是偶爾提及:拼湊出成果的,不只是技術,而是反覆試驗和一點點不甘心。
面對解析度不足和資料管理難題,我們該如何迎接未來的挑戰?
大多數研究人員面對活體成像解析度不夠清楚、資料暴增難管理,還有AI模型在不同場景下表現飄忽不定這幾個困擾,好像都在找解法卻沒一條路完全打通。其實市面上已經有人試著把多中心影像資料丟到雲端協作平台裡,偶爾也會搭配分層權限和加密措施,讓跨單位討論比較順暢。至於螢光訊號容易淬滅的問題,有團隊會換用新型標記或調整曝光週期,但是否每種細胞環境都適用,目前看起來還要再觀察。資料標註這塊,有人嘗試半自動化標記流程,減輕判讀壓力,但人工校正還是跑不掉一部分。最後,如果臨時碰到特殊樣本異常,不少實驗室現在傾向建立快速回饋小組,必要時直接改變操作步驟或重設參數,大概能減少延誤。不過每個方案都有局限,要挑哪個方法還是得根據現場需求和資源來衡量。


















































